Abstract
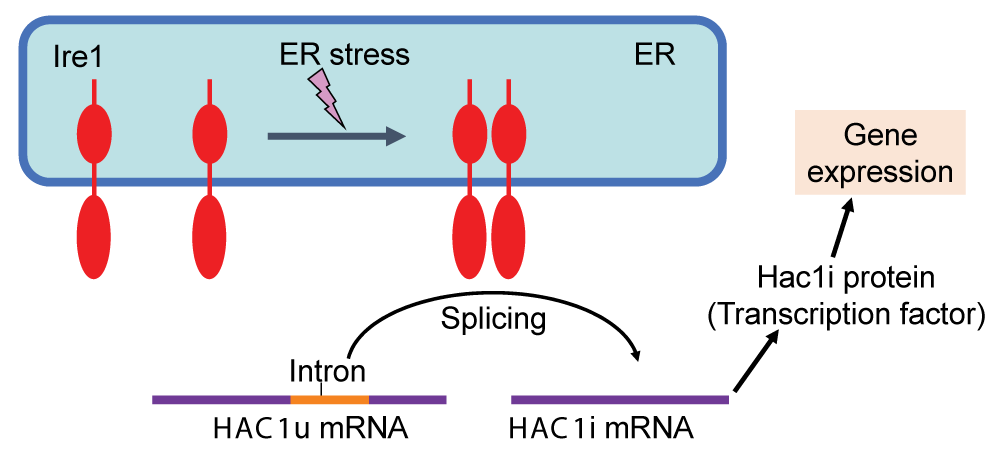
Upon dysfunction of the Endoplasmic Reticulum (ER), eukaryotic cells provoke a gene expression program, namely, the Unfolded Protein Response (UPR), leading to an increase in the size and function of the ER. In the yeast Saccharomyces cerevisiae, the UPR is modulated by the Hac1i protein, which is a transcription factor produced by ER stress. When the UPR is artificially triggered under non-stress conditions by artificial expression of the Hac1i protein, S. cerevisiae cells carry an enforced and enlarged ER, which allows us to obtain commercially valuable materials such as secretory proteins and functional lipids abundantly.




![Fluorescence microscopic analysis of S. cerevisiae cells Wild-type S. cerevisiae cells (BY4742 (MATα, ura3, leu2, his3, lys2): WT cell) and their derivative carrying the HAC1i expression plasmid (Hac1i cell) were cultured in yeast standard synthetic complete (SC) medium at 30 °C and fluorescence microscopically observed. To generate the HAC1i expression plasmid, the HAC1i gene was cloned into the Tet-off vector pCM190 [20], leading to inducible expression of the Hac1i protein upon culturing cells in the SC medium. (A) Cells were stained with BODIPY 493/503 to visualize neutral lipids. Hac1i cells carried larger and more abundant lipid droplets than WT cells. (B) The fluorescent ER marker protein, Elo2-mCherry, was expressed from a plasmid that had been created by insertion of the ELO2 gene, which encodes an ER membrane protein Elo2, into the mCherry expression plasmid pYT-TDH3p-PMA1-mCherry [21]. The ER in Hac1i cells was more expanded than in WT cells. Red dashed lines represent the cell outline.](https://www.igminresearch.com/articles/figures/igmin142/igmin142.g002.png)



